
University of Geneva - Faculty of Medicine

Others Links
Faculty of Medicine
CMU-Library
Medline-Pubmed
Host Pathogens Interactions
Home
| The Linder Lab | |||
|---|---|---|---|
|
 |
Pr. P. LINDER CMU/ Dpt MIMOL Rue Michel-Servet 1 1211 Genève 4 Suisse Patrick.Linder@unige.ch Tel.: 022 379 54 84 Fax: 022 379 55 02 |
|
| Research's subject | Group's publications | Team's members | To Find |
| _________________________________________________________________ | |||
| Research's subject | |||
| Acquisition and expression of virulence factors in Staphylococcus aureus. | |||
Bacteria are everywhere and life without them is not possible. Most bacteria are harmless to human being, some are obligatory pathogens where their presence is equivalent to disease, others may not necessarily cause disease. Amongst this later category is the opportunistic pathogen Staphylococcus aureus. This Gram-positive bacterium forms golden grapes and is present in the nose of 30-50% of the people. In most of the healthy carriers, the presence of this bacterium is not noticeable, but in certain cases it can cause diseases ranging from furuncles to life threatening systemic infections. S. aureus is regularly in the Headlines because of its multiple resistances to antibiotics, including methicilin (MRSA, methicilin resistant S. aureus). Persistent infections of S. aureus are at least in part due to its extraordinary capability to form biofilms and to survive intracellularly. . 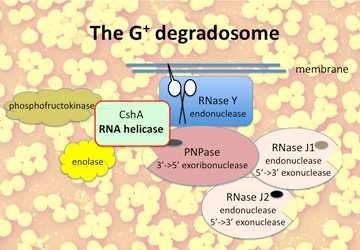 Our laboratory has a long-standing interest in RNA helicases of the DEAD-box protein family. In eukaryotic cells, DEAD-box RNA helicases are involved in processes as divers as transcription, pre-mRNA splicing, ribosome biogenesis, RNA export, translation initiation, NMD (non-sense mediated decay), and organelle gene expression. All these processes are dynamic ribo-nucleo-protein (RNP) particles that need to be assembled, undergo conformational changes, and eventually be disassembled. Indeed a wonderful playground for RNA helicases! Our laboratory has a long-standing interest in RNA helicases of the DEAD-box protein family. In eukaryotic cells, DEAD-box RNA helicases are involved in processes as divers as transcription, pre-mRNA splicing, ribosome biogenesis, RNA export, translation initiation, NMD (non-sense mediated decay), and organelle gene expression. All these processes are dynamic ribo-nucleo-protein (RNP) particles that need to be assembled, undergo conformational changes, and eventually be disassembled. Indeed a wonderful playground for RNA helicases!The DEAD-box proteins are characterised by the presence of 12 highly conserved amino acid motifs that are involved in the binding of the substrates, ATP and RNA, and in intramolecular interactions. Unwinding of double stranded RNA or the dissociation of proteins from RNA does require ATP binding and ultimately ATP hydrolysis. Although not as abundant as in eukaryotes, most bacteria encode RNA helicases of the DEAD-box protein family. They have been identified by genetic screens or by bio-informatic analyses. In contrast to most eukaryotic DEAD-box protein genes, they are not essential under 'normal' (laboratory) growth conditions. However, even subtle differences in growth rate may result in huge differences in population density because of the short generation time of bacterial growth. Moreover, bacteria encounter various growth conditions that may require DEAD-box proteins. Indeed, early in the discovery of DEAD-box proteins, they were found to be associated with cold-shock in Escherichia coli. The S. aureus genome encodes two DEAD-box RNA helicases. One of them was identified in a screen for biofilm deficient mutants in a clinical strain. Although growth of this mutant strain at temperatures of 37°C and above is normal, this strain is highly deficient in biofilm formation. At present we are analysing the role of this RNA helicase in gene expression to elucidate its role in biofilm formation (collaboration with the laboratory of Jacques Schrenzel, Genomic Research Laboratory, University Hospital, Geneva). In a parallel project we are currently analysing the second RNA helicase encoded by the S. aureus genome. Disruption of this gene does confer a cold sensitive phenotype, but the strain grows normally at 37°C. Other projects in the laboratory are related to RNA metabolism in general. Last but not least, we are trying to develop new strategies to make clinical S. aureus strains amenable to molecular genetic methods. Indeed, many of the clinical strains used in our studies cannot be transformed with plasmid DNA, even if the plasmids were prepared in the permissive laboratory strain RN4220. In this work we inactivate the host mediated restriction by the insertion of group II introns. |
|||